Welcome to metaGOflow’s documentation!
metaGOflow is a
Common Workflow Language (CWL)
based pipeline
for the analysis of shotgun metagenomic data at the sample level.
It was initially built to address the needs of the
EMO BON community
but it can be used for any type of shotgun sequencing data.
metaGOflow is based on the tools and subworkflow implemented in the
framework of MGnify.
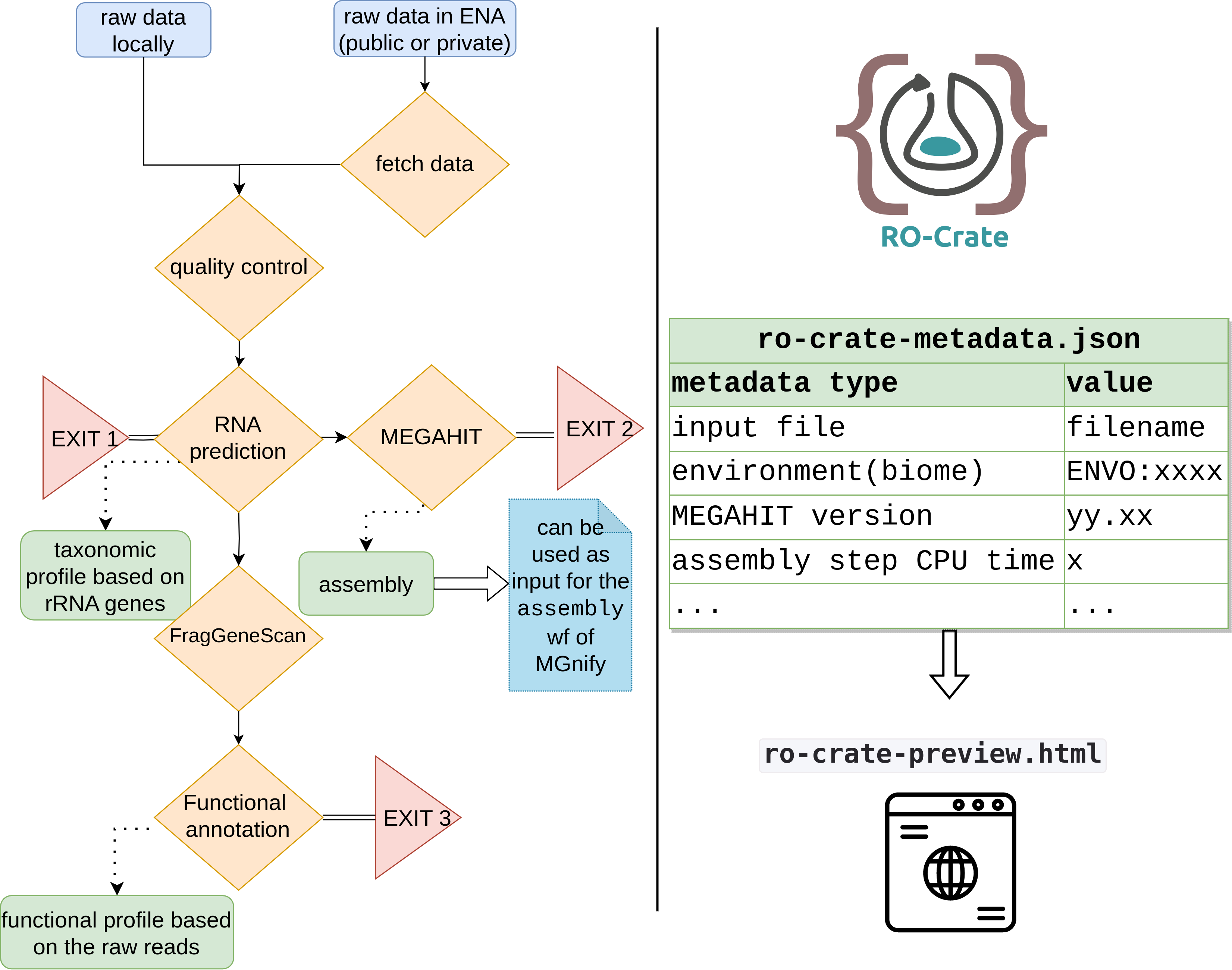
It produces a Research Object Crate (RO-Crate) as its main output that includes the taxonomic inventory and/or the functional profile of the sample based on the user’s settings.
Check out the How to run metaGOflow section for further information, including Installation instructions for the project.
Note
If you find metaGOflow useful, remember to cite it:
Haris Zafeiropoulos, Martin Beracochea, Stelios Ninidakis, Katrina Exter, Antonis Potirakis, Gianluca De Moro, Lorna Richardson, Erwan Corre, João Machado, Evangelos Pafilis, Georgios Kotoulas, Ioulia Santi, Robert D Finn, Cymon J Cox, Christina Pavloudi, metaGOflow: a workflow for the analysis of marine Genomic Observatories shotgun metagenomics data, GigaScience, Volume 12, 2023, giad078, 10.1093/gigascience/giad078.
Contents

